Tet3-mediated DNA Hydroxylation is Involved in Epigenetic Reprogramming by Oocytes
Source:
Time: 2011-09-07
Sperm and eggs carry distinctive epigenetic modifications that are adjusted by reprogramming following fertilization. The paternal genome in a zygote undergoes active DNA demethylation before the first mitosis. The biological significance and mechanisms of this paternal epigenome remodeling have remained unclear.
A team of researchers, led by Dr. XU Guoliang and Dr. LI Jinsong from the Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Sciences, CAS, reported that, within mouse zygotes, oxidation of 5-methylcytosine (5mC) occurs on the paternal genome, changing 5mC into 5-hydroxymethylcytosine (5hmC), and the maternal Tet3 is required for this conversion. Deletion of Tet3 from oocytes impedes the demethylation process of the paternal Oct4 and Nanog genes. The heterozygous mutant offspring lacking maternal Tet3 suffer an increased incidence of developmental failure. Furthermore, Oocytes lacking Tet3 also appear to have reduced ability to reprogram the injected nuclei from somatic cells. These results suggest that Tet3-mediated DNA hydroxylation is involved in epigenetic reprogramming of the zygotic paternal DNA following natural fertilization and may also contribute to somatic cell nuclear reprogramming during animal cloning.
This work entitled “The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes” was published online in Nature on September 4th, 2011.
This study was supported by the grants from the Chinese Academy of Sciences, the Ministry of Science and Technology, and National Natural Science Foundation of China..
AUTHOR CONTACT:
XU Guoliang
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
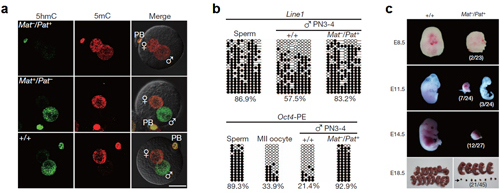
The role of Tet3 in 5mC oxidation, demethylation of paternal DNA, and embryonic development
a. 5hmC (green) and 5mC (red) immunostaining in wild-type (+/+) and maternally (Mat−/Pat+) or paternally (Mat+/Pat−) Tet3-deficient zygotes at the PN5 stage.
b. Methylation analysis of Line1 and Oct4 (PE, the proximal enhancer region) in male pronuclei isolated from wild-type (+/+) and Tet3-deficient (Mat−/Pat+) zygotes. Open and filled circles represent unmethylated and methylated CpG sites, respectively.
c. Developmental failure among embryos lacking maternal Tet3. (Image provided by Dr. XU Guoliang)
 Appendix:
Appendix: