Researchers Reveal Translational Quality Control Mechanism Mediated by Yeast LeuRS
Source:
Time: 2013-08-30
Aminoacyl-tRNA synthetases should ensure high accuracy in tRNA aminoacylation. However, the structural similarity between amino acids is always a challenge for some aaRSs, such as leucyl-tRNA synthetase (LeuRS), which require editing function to remove mis-activated amino acids. Recently, scientists from Chinese Academy of Sciences (CAS) revealed aminoacylation and translational quality control strategy employed by LeuRS from
Candida albicans cytoplasm (
CaLeuRS) with genetic code ambiguity.
About half of the aaRSs misactivate non-cognate amino acids and mischarge their cognate tRNA with the non-cognate amino acids. To solve this problem, the proofreading (editing) mechanism of aaRS has evolved to hydrolyze the mis-intermediate and mis-products. Editing can occur at the aa-AMP level (pre-transfer editing) and/or mis-aminoacyl-tRNA level (post-transfer editing), depending on the various aaRSs. Furthermore, in the cytoplasm of the human pathogen
Candida albicans, the CUG codon is translated as both Ser and Leu by a uniquely evolved
CatRNA
Ser (CAG).
CaLeuRS is a crucial component for CUG codon ambiguity and harbors only one CUG codon at position 919. Dr. ZHOU Xiaolong and his colleagues under the guidance of Prof. WANG Enduo at Shanghai Institute of Biochemistry and Cell Biology, CAS, found that LeuRSs from yeast have a relaxed tRNA recognition capacity. Comparison of
CaLeuRS-Ser
919 and
CaLeuRS-Leu
919 revealed potential regulating effect of genetic code alteration on protein function. The mis-activation and editing of non-cognate amino acids by
CaLeuRS were studied. Interestingly, they found that
CaLeuRS is naturally deficient in tRNA-dependent pre-transfer editing for non-cognate Nva while displaying a weak tRNA-dependent pre-transfer editing capacity for non-cognate ABA. They also showed that post-transfer editing of
CaLeuRS is not tRNA
Leu species-specific. In addition, other eukaryotic but not archaeal or bacterial LeuRSs were found to recognize
CatRNA
Ser (CAG).
This work systematically studied the aminoacylation and editing properties of
CaLeuRS and established a characteristic LeuRS model with naturally deficient tRNA-dependent pre-transfer editing, which increases LeuRS types with unique editing patterns. It improves our understanding about the mechanism of genetic code ambiguity of
C. albicans and opened the window for modifying its genetic code character and potentially changing its proteome and diseasing-causing characteristics. In addition, the capacity of eukaryotic LeuRSs at aminoacylating
CatRNA
Ser suggests the possibility of reconstructing proteome of other eukaryotes by simply introducing this unique tRNA
Ser.
This study entitled “
Aminoacylation and translational quality control strategy employed by leucyl-tRNA synthetase from a human pathogen with genetic code ambiguity” has been published online in
Nucleic Acids Research on Aug 23
rd, 2013. It is funded by the National Natural Science Foundation of China, the National Key Basic Research Foundation of China, the Chinese Academy of Sciences and Committee of Science and Technology in Shanghai.
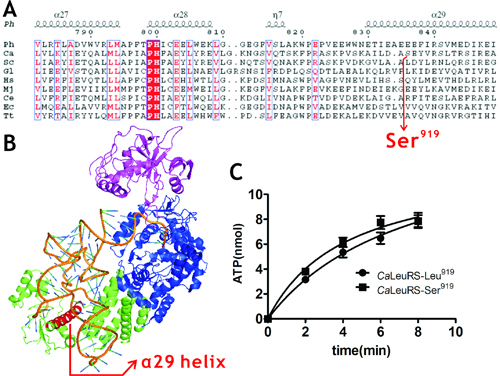
Location of residue 919 and its effect on amino acid activation in CaLeuRS. (Image by Prof. WANG Enduo’s group)

 Appendix:
Appendix: