Scientists Reveal New Insights into The Fanconi Anemia Pathway
Source:
Time: 2013-09-17
The Fanconi anemia (FA) pathway plays a critical role in the repairing of DNA interstrand cross-links (ICLs) during S-phase. Recently, scientists from Chinese Academy of Sciences (CAS) determined the crystal structure of the FANCM-FAAP24 complex and provided new insights into its functions in DNA repair.
The central event of the FA pathway is the monoubiquitination of the FANCI-FANCD2 complex, which is mediated by the FA core complex consisting of eight FA proteins and five FA-associated proteins. FANCM is a key component of the FA core complex, which can interact with FAAP24 (FA-associated protein 24 kDa) and the MHF1-MHF2 (FANCM-associated histone-fold proteins 1 and 2) complex via different regions. The FANCM-FAAP24 complex binds preferentially to single-strand DNA and plays key roles in recognizing the DNA ICLs, recruiting the FA core complex, and initiating ATR (ataxia-telangiectasia and Rad3-related)-mediated checkpoint signaling. The FANCM-MHF complex binds preferentially to branched DNA and double-strand DNA and stimulates the DNA branch migration and replication fork reversal activities of FANCM.
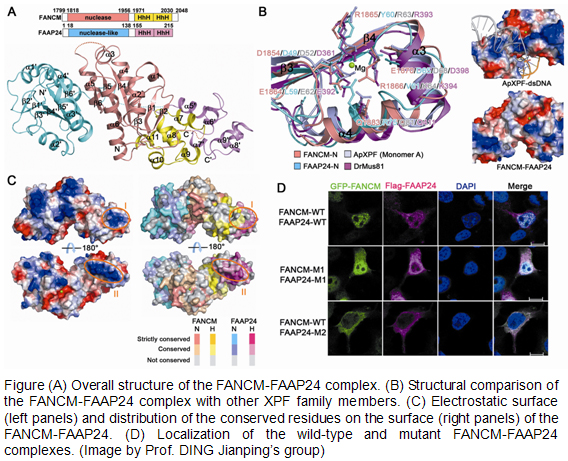
Both FANCM and FAAP24 belong to the XPF family, which contains members functioning as structure-specific endonucleases. However, the FANCM-FAAP24 complex has no detectable nuclease activity. The molecular basis for the nuclease inactivity remains elusive. The recognition of DNA lesion by the FANCM-FAAP24 complex is unknown. Now, PhD student YANG Hui and her colleagues, led by Prof. DING Jianping, from the Shanghai Institute of Biochemistry and Cell Biology, CAS, determined the crystal structure of the C-terminal segment of FANCM in complex with FAAP24.
Structural comparisons with the catalytic members of the XPF family together with biochemical data revealed that the variation of key residues at the active site and the DNA-binding incapability of the surrounding region account for the lack of nuclease activity of the FANCM-FAAP24 complex. Based on structural analyses, they identified a potential DNA-binding site in particularly for ssDNA in the FANCM-FAAP24 complex which was further verified by immunofluorescence and electrophoretic mobility shift assays. They concluded that the first HhH motif of FAAP24 is essential for targeting the FANCM-FAAP24 complex to chromatin. The two regions of FANCM interacting with MHF and FAAP24 might be located in proximity in the three-dimensional structure, such that MHF and FAAP24 could bind to dsDNA and ssDNA regions of one stalled replication fork, respectively, with high affinity and then cooperate to anchor FANCM to the branch point of the replication fork. These results provide new insights into the molecular basis for the functions of the FANCM-FAAP24 complex in the FA pathway in DNA damage repairing.
This study entitled “
Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair” was published online in
Nucleic Acids Research on September 3
rd, 2013. The work was supported by grants from the Ministry of Science and Technology of China and the National Natural Science Foundation of China.

 Appendix:
Appendix: