Aminoacyl-tRNA synthetases catalyze the first step of protein synthesis, aminoacylation of tRNA, and provide correct building blocks for ribosomal protein translation. Rate and accuracy of aminoacylation, established by tRNA charging and editing, determine the efficiency and fidelity of protein translation.
In majority species, mitochondrial genome contains two genes for tRNALeu, tRNALeu (CUN) (N: A, C, G, T) and tRNALeu(UUR) (R: A, G) and one tRNAThr gene. However, in some yeasts, tRNALeu(CUN) gene is lost a very unique tRNAThr (tRNAThr1) is evolved. This tRNAThr1 is a very specific example of ususual 8-nt tRNA anti-codon and reassigns CUN codon from Leu to Thr. Moreover, yeast mitochondrial threonyl-tRNA synthetase (ScmtThrRS) contains only aminoacylation and tRNA binding domains but naturally lacks editing domain.
Recently, Dr. ZHOU Xiaolong and his colleagues led by Professor WANG Enduo at Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, CAS showed that ScmtThrRS mis-activates non-cognate Ser and harbors tRNA isoacceptor-specific tRNA dependent pre-transfer editing capacity. Indeed, tRNAThr2 but not tRNAThr1 stimulates significant pre-transfer editing. Actually, intrinsic ability of tRNAThr1 to trigger pre-transfer editing requires the presence of editing domain. This is the first example of tRNA synthetase exhibiting such a isoacceptor specificity during proofreading. Finally, a yeast MST1 gene knockout strain is constructed and in vivo complementation assays show that the catalytic core and tRNA binding domains of ScmtThrRS co-evolve to recognize the unusual tRNAThr1. In combination, the results provide evidences showing that tRNA-dependent pre-transfer editing takes place in the aminoacylation catalytic core.
This work entitled “A minimalist mitochondrial threonyl-tRNA synthetase exhibits tRNA-isoacceptor specificity during proofreading” is published online in Nucleic Acids Research on Nov 20, 2014, and is funded by the Natural Science Foundation of China and the National Key Basic Research Foundation of China.
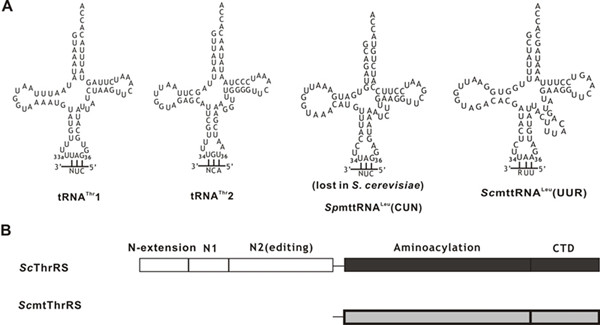
Figure. Representations of tRNAs and S. cerevisiae ThrRSs investigated in this study.
(A) Cloverleaf structures of S. cerevisiae mitochondrial tRNAThr1, tRNAThr2, tRNALeu(UUR) [ScmttRNALeu(UUR)] and S. pombe mitochondrial tRNALeu(CUN) [SpmttRNALeu(CUN)], which has been lost in S. cerevisiae mitochondria during evolution. (B) Linear representation of the domain arrangement of SccytThrRS and ScmtThrRS. Aminoacylation domain and C-terminal tRNA binding domain (CTD) of SccytThrRS or ScmtThrRS are colored in black or gray, respectively.
(Image provided by Prof. WANG Enduo’s group.)
 Appendix:
Appendix: