A complex disease is generally a problem resulted from the failure of the relevant system, and thus should be investigated in a dynamic and network manner. Therefore, compared with single molecules, networks or edges among molecules are considered to be a stable and reliable form for characterizing complex diseases or phenotypes as biomarkers. Generally, an edge in a molecular network is represented by its correlation coefficient which requires several samples to calculate. But for many biomedical applications, there is only one sample for one individual. So it is difficult to diagnose an individual with a single sample by using correlation coefficient.
To address this issue, Prof. CHEN Luonan and his group from the Key Laboratory of Systems Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences of CAS developed a new method for diagnosing single-sample individual by using the correlation information of molecular pairs, i.e. edge biomarkers. In this method they designed a data transformation that turns ‘node data’ of molecular expression into ‘edge data’ which capture the information of correlation for each sample. Then feature selection methods are applied to edge data to extract the features with best classification accuracy as edge biomarkers.
They applied the method on lung adenocarcinoma dataset from The Cancer Genome Atlas (TCGA) database and showed the advantages of edge biomarkers: (i) they achieved better cross-validation accuracy of diagnosis than molecule or node biomarkers, (ii) edge biomarkers are significantly enriched in relevant biological functions or pathways, implying that the association changes in a network, rather than expression changes in individual molecules, tend to be causally related to cancer development, (iii) our method identifies edge biomarkers even with non-differential molecules, which are generally missed by traditional works.
This work entitled “Diagnosing phenotypes of single-sample individuals by edge biomarkers” was published in Journal of Molecular Cell Biology on April 26, 2015. This work was supported by the grants from the Chinese Academy of Sciences and the National Natural Science Foundation of China.
CONTACT:
CHEN Luonan, Principal Investigator
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
Shanghai 200031, China.
E-mail: lnchen@sibs.ac.cn
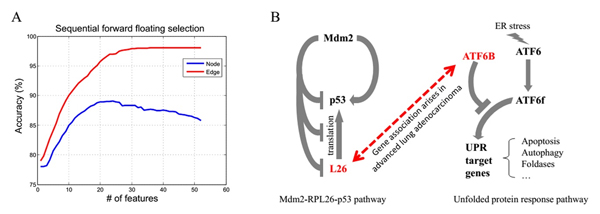
Fig. (A) Applying feature selection method to node and edge data shows that edge features achieved better cross-validation accuracy. (B) A hypothesis that the association between the expressions of RPL26 and ATF6B is related to the development of lung adenocarcinoma from earlier stage into advanced stage is proposed.
(Image provided by Prof. CHEN Luonan’s group)
 Appendix:
Appendix: