Disease genes are key elements to study or treat diseases, and their altered regulations or interactions in complex diseases are even more important than the expression variety of genes themselves, revealing the abnormal cooperation among multiple genes. Many laboratories have studied the same diseases, but there are several unavoidable issues to detect consistent disease genes or interactions from these studies: (i) the experiment design, platform, data analysis, or final conclusions on a disease would be not the same due to different research topics and preferences of individual researchers; (ii) the accumulated high-throughput data usually have unbalanced information, e.g., less control samples than case samples.
To overcome above problems, Prof. CHEN Luonan and his group from the Key Laboratory of Systems Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, CAS developed novel mathematical model of consistently differential interaction (CDI) and CDI-based method to detect disease genes of a disease by integrating multiple independent datasets. The assumption underlying CDI-based methods is that, if the interactions or regulations are always identified from independent disease datasets, these abnormal behaviors, e.g., regulations or interactions, are considered to be strongly related to the disease and would be disease-related edges.
Researchers applied CDI-based method on the data of non-small cell lung cancer (NSCLC) from TCGA, including lung adenocarcinoma (LUAD) and lung squamous cell cancer (LUSC). The obtained CDIs were significantly enriched with cancer genes, especially lung cancer-specific genes. Moreover, it also has been shown that those differential interactions have better subtype specificity than differential expressed genes, and they can be directly used as new edgetic targets from the network viewpoint, both to accurately diagnose the disease phenotypes and to predict patient overall survival. Finally, a potential molecule mechanism has been derived as that the gene markers or gene-pair markers regulate the expressions or interactions of known metastasis factors of lung cancer, thereby enhancing the metastatic process.
This work entitled "Detecting disease genes of non-small lung cancer based on consistently differential interactions" was published in Cancer and Metastasis Reviews on May 26, 2015. This work was supported by the grants from the Chinese Academy of Sciences and the National Natural Science Foundation of China.
CONTACT:
CHEN Luonan, Principal Investigator
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
Shanghai 200031, China.
E-mail: lnchen@sibs.ac.cn
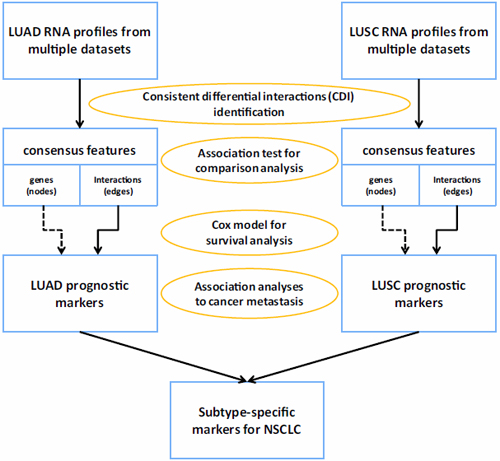
Fig. 1 Overview of framework corresponding to CDI-based method. The framework has main four steps to obtain reliable subtype-specific markers for NSCLC.
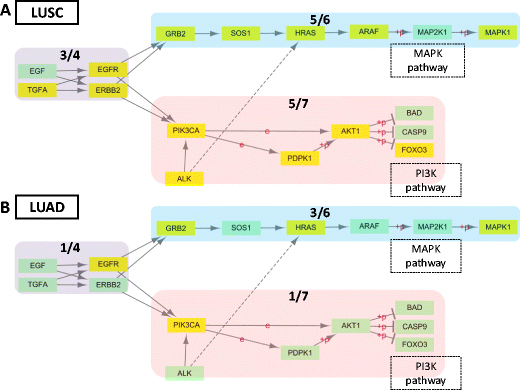
Fig. 2 The predicted genes by CDI-based method are enriched in non-small cell lung cancer pathway. The predicted gene sets are enriched in NSCLC pathway for LUSC (A) and for LUAD (B). The identified gene sets by CDI-based method are colored yellow in pathway.
(Image provided by Porf. CHEN Luonan’s group)
 Appendix:
Appendix: