tRNA is one of the main RNA moleculars in cell, and tRNAs play central roles in protein synthesis by transfer amino acids to ribosome. Nucleotides in tRNA are often post-transcriptionally modified, especially residues from the anti-codon region, some modifications are essential for the function of tRNA and lack of these modifications can cause some severe cellular deficiency in vivo. TrmL is the prokaryotic methyltransferase which is responsible for the transfer of the methyl group from S-adenosyl-L-methionine to the wobble base of tRNALeuCAA and tRNALeuUAA isoacceptors. TrmL is one of the minimalist SPOUT methyltransferase, the tRNA recognition determinant by TrmL remains elusive.
Under the guidance of Prof. WANG Enduo and Dr. LIU Rujuan, Ph.D students ZHOU Mi et al identified the tRNA recognition elements by TrmL through site directed mutagenesis on tRNAs. By measuring tRNA methylation activity and assaying kinetic parameters of tRNA mutants, they found that TrmL exhibits a fine-tuned tRNA substrate recognition mechanism. The results show that TrmL could recognize RNA substrate through the stem-loop structure but the L-shape tertiary structure of tRNA is not necessary. TrmL may stretch out many hands and arms, to identify those recognition determinants at the same time. Specifically, those recognition determinants include the correct identity elements of the anticodon loop, the structure of anticodon stem and loop, a pyrimidine at the wobble position, and a preexisting i6A37 modification.
Methylation of RNA is one of the most common and ubiquitous modifications involved in many important life processes. This work contributes to the understanding of tRNA methylation interdependence for biosynthesis and enzyme recognition, both of which are ongoing topics of interest to the field.
This work was published in RNA Biology entiled “Identification of determinants for tRNA substrate recognition byEscherichia coliC/U34 2’-O-methyltransferase” on Jun 24th, 2015. This work is funded by the National Natural Science Foundation of China, the Ministry of Science and Technology of China, the Chinese Academy of Sciences and Science and Technology Commission of Shanghai Municipality.
CONTACT:
WANG Enduo, Principal Investigator
Institute of Biochemistry and Cell Biology, Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences
Shanghai 200031, P. R. China
Phone: 86-21-54921241
E-mail: edwang@sibcb.ac.cn
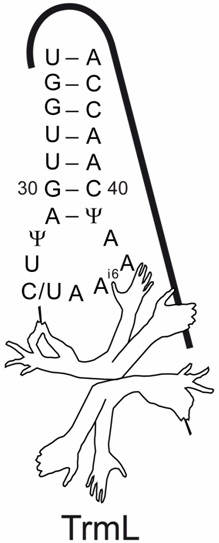
Fig. A scheme to summarize the determinant elements of tRNA recognized by TrmL.
(Image provided by Prof. WANG Enduo’s group)
 Appendix:
Appendix: