Researchers Reveal the Catalytic Mechanism of Isoorotate Decarboxylase
Source:
Time: 2015-09-20
TET proteins participate in the active DNA demethylation and the product 5-carboxyl-cytosine could be converted to cytosine through two possible mechanisms. One possibility is that a putative DNA decarboxylase catalyzes the direct decarboxylation of 5-carboxyl-cytosine to cytosine. Now, researchers from Chinese Academy of Sciences (CAS) reveal the catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed lights on the search for DNA decarboxylase.
Methylation of cytosine in DNA is an important epigenetic modification which plays crucial roles in biological processes. It is well established that DNA methylation is catalyzed by DNA methyltransferases. However, the issue about how active DNA demethylation occurs has been in controversial for years. Currently, it is believed that the TET proteins in mammals are involved in the process by stepwise oxidizing 5-methyl-cytosine to 5-carboxyl-cytosine which can be further converted to cytosine via two possible mechanisms. One possibility is that 5-carboxyl-cotysine can be directly decarboxylated by a putative DNA decarboxylase. The chemistry for the conversion of 5-methyl-cytosine to 5-carboxyl-cytosine and possibly to cytosine in the active DNA demethylation in mammals is very similar to that for the conversion of thymidine to 5-carboxyl-uracil and further to uracil in fungi. Thus, it is believed that a putative DNA decarboxylase may share some similarities in sequence, structure and catalytic mechanism with Isoorotate Decarboxylase (IDCase). Besides, the substrate recognition and catalytic mechanism of IDCase remain elusive.
Recently, PhD students XU Shu-tong and LI Wen-jing and their colleagues, led by Prof. DING Jianping form the Shanghai Institute of Biochemistry and Cell Biology (SIBCB), CAS, solved the crystal structures of the wild-type and mutant
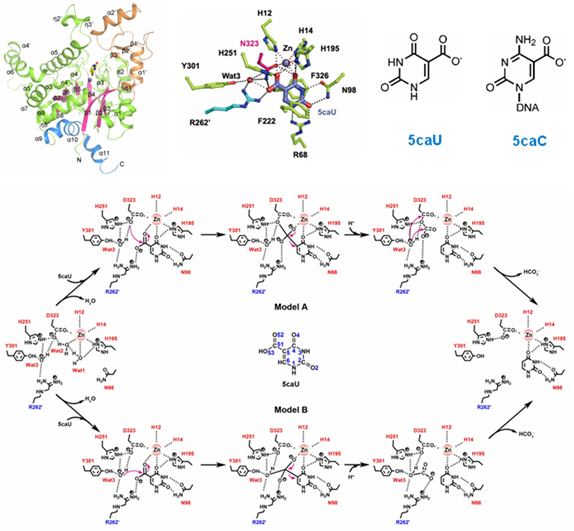
Cordyceps militaris IDCase in apo form and in complexes with the substrate 5-carboxyl-uracil, a substrate analog 5-nitro-uracil, and the product uracil, and the wild-type
Metarhizium anisopliae IDCase in apo form. The functional roles of the key residues involved in interactions with Zn
2+ and the substrate are validated by mutagenesis and biochemical studies. The results showed that IDCases belong to the amidohydrolase superfamily and function as dimmers. The structural and biochemical data together revealed the molecular bases of the substrate recognition and binding and a novel catalytic mechanism of decarboxylation for IDCases. In addition, these results provide useful hints for the search of potential DNA decarboxylase in mammals.
This research entitled “
Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase” was published online in
Cell Research on Aug 6
th, 2013. This study was helped by Prof. XU Guo-liang from SIBCB. This study was supported by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China, and the Science and Technology Commission of Shanghai Municipality.
IDCase adopts the typical (β/α)8 barrel fold of the amidohydrolase superfamily, the key residues involved in the binding of the metal ion, the substrate, and the catalysis are identified, and a novel catalytic mechanism of IDCase is proposed (Image by Prof. DING Jianping’s group).
 Appendix:
Appendix: