The epigenomes of mammalian sperm and oocytes, characterized by gamete-specific 5-methylcytosine (5mC) patterns, are reprogrammed during early embryogenesis to establish full developmental potential. Previous studies have suggested that the paternal genome is actively demethylated in the zygote while the maternal genome undergoes subsequent passive demethylation via DNA replication during cleavage. Active demethylation is known to depend on 5mC oxidation by Tet dioxygenases and excision of oxidized bases 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by thymine DNA glycosylase (TDG). However, whether Tet-TDG mediated oxidative demethylation plays a role in the zygote is unclear.
A research team led by Prof. XU Guoliang and Prof. LI Jinsong, at Shanghai Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences , in collaboration with Prof. Tang Fuchou, at the Biodynamic Optical Imaging Center, Peking University, demonstrate the role of active and passive demethylation directly by looking at maternal and paternal pronuclear DNA. Taking advantage of mouse models characterized by germline-specific knockout of Tet3 and Tdg and the improved reduced representation bisulfite sequencing (RRBS) approach, the team determined the methylomes at base resolution on a genome-wide scale. They further validated the modification states of selected genomic loci using the hairpin-bisulfite analysis and the newly developed M.SssI-assisted bisulfite (MAB) sequencing method which is allowed to distinguish cytosine from its modified forms.
The study showed that, both maternal and paternal genomes undergo widespread active and passive demethylation in zygotes before the first mitotic division. Passive demethylation was blocked by the replication inhibitor aphidicolin, and active demethylation was abrogated by deletion of Tet3. At actively demethylated loci, 5mCs were processed to unmodified cytosines but higher oxidation intermediates 5fC and 5caC were not detected. Surprisingly, the demethylation process was unaffected by the deletion of TDG from the zygote, suggesting the existence of other demethylation mechanism downstream of Tet3-mediated oxidation.
The work above entitled “Active and Passive Demethylation of Male and Female Pronuclear DNA in the Mammalian Zygote” was published in Cell Stem Cell on Oct 2, 2014. Research funding came from the Ministry of Sciences and Technology of China, the National Natural Science Foundation of China, the “Strategic Priority Research Program” of the Chinese Academy of Sciences and the National Science and Technology Major Project of China.
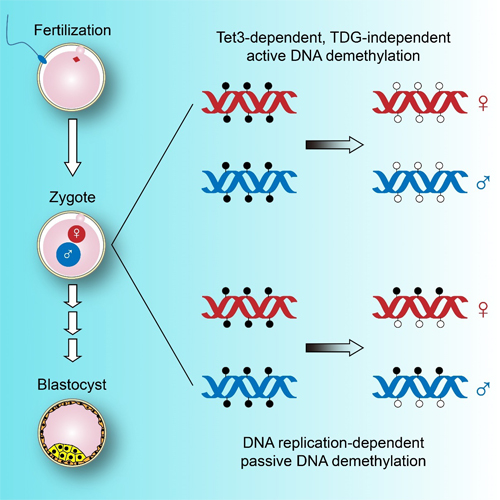
In one-cell mouse embryos, the maternal and paternal genomes both undergo Tet3-dependent active demethylation and replication-mediated passive demethylation. Surprisingly, the active demethylation pathway does not require TDG. (Image provided by Prof XU Guoliang`s group)
 Appendix:
Appendix: