The interaction mechanism between leucyl-tRNA synthetases and tRNALeu in haloalkaliphile
Source:
Time: 2015-09-20
On Feb 6, 2014,
Nucleic Acids Research online published the work entitled “
Coexistence of bacterial leucyl-tRNA synthetases with archaeal tRNA binding domains that distinguish tRNALeuin the archaeal mode” from Prof. Enduo Wang’s group in Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.
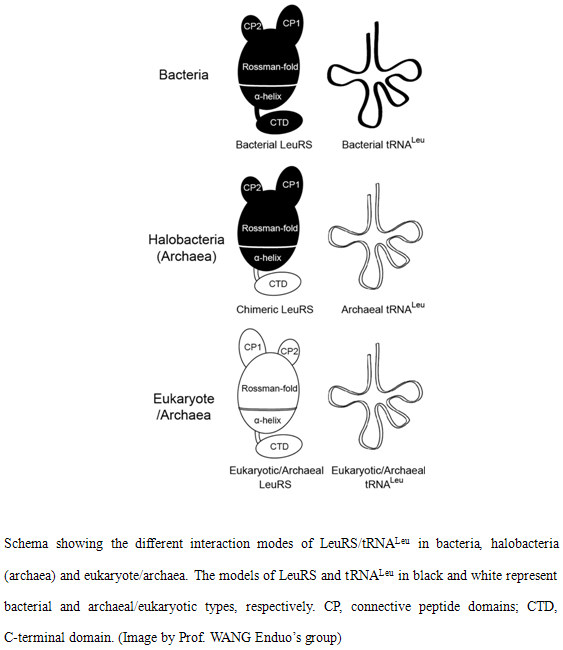
Leucyl-tRNA synthetase (LeuRS) is a multi-domain enzyme, which is divided into bacterial and archaeal/eukaryotic types. In general, one specific LeuRS, the domains of which are of the same type, exists in a single cell compartment. However, some species, such as the haloalkaliphile
Natrialba magadii, encode two cytoplasmic LeuRSs,
NmLeuRS1 and
NmLeuRS2, which are the first examples of naturally occurring chimeric enzymes with different domains of bacterial and archaeal types. Furthermore,
N. magadii encodes typical archaeal tRNA
Leus. The tRNA recognition mode, aminoacylation and translational quality control activities of these two LeuRSs are interesting questions to be addressed.
FANG Zhi-Peng and his colleagues guided by Professor WANG En-Duo and Associate Professor ZHOU Xiao-Long successfully purified active
NmLeuRS1 and
NmLeuRS2 after gene expression in
E. coli. They discovered that
NmLeuRSs distinguished cognate
NmtRNA
Leu in the archaeal mode, whereas the N-terminal region was of the bacterial type. However,
NmLeuRS1 exhibited much higher aminoacylation and editing activity than
NmLeuRS2, suggesting that
NmLeuRS1 is more likely to generate Leu-tRNA
Leu for protein biosynthesis. Moreover, using
NmLeuRS1 as a model, they demonstrated mis-activation of several non-cognate amino acids, and accuracy of protein synthesis was maintained mainly via post-transfer editing. This comprehensive study of the
NmLeuRS/tRNA
Leu system provides a detailed understanding of the coevolution of aaRSs and tRNA, and the mechanism of aminoacyl-tRNA synthetase on quality control of protein biosynthesis in the haloalkaliphilic bacterium.
This study was supported by the National Basic Research Program of China, the Natural Science Foundation of China and the Chinese Academy of Sciences.
AUTHOR CONTACT:
WANG En-Duo
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, the Chinese Academy of Sciences, Shanghai, China
Phone: 86-21-5492 1241;
E-mail: edwang@sibcb.ac.cn

 Appendix:
Appendix: