Studies Shed Light on the Evolutionary Origin of Key Component of Adaptive Immune System
Source:
Time: 2015-09-20
Many hypotheses have been proposed regarding the origin and evolution of recombination activating genes (RAG), which is responsible for generating a highly diversified repertoire of antigen receptors in the vertebrate immune system. It is not clear why adaptive immune system is restricted to the genomes of jawed vertebrates, and no functionally similar homologs of RAG had ever been found from animals outside of the jawed vertebrates.
A research team led by Prof. LIU Xiaolong from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences showed that an RAG1-like DNA fragment (bfRAG1L) encodes a protein resembling an ancient form of RAG1.
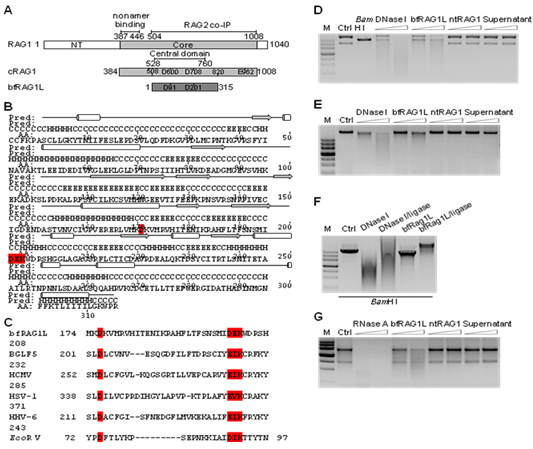
They showed that, in amphioxus genome, an ancient RAG1-like DNA fragment (bfRAG1L) that encodes a virus-related protein resembling the central domain of core RAG1 (cRAG1). The bfRAG1L contains an unexpected retroviral type II nuclease active site motif, DXN(D/E)XK, and is capable of degrading both DNA and RNA. Moreover, bfRAG1L shares important functional properties with the central domain of cRAG1, including interaction with RAG2 and localization to the nucleus. Remarkably, the reconstitution of bfRAG1L into a cRAG1-like protein yielded an enzyme capable of recognizing recombination signal sequences (RSSs) and performing V(D)J recombination in the presence of mouse RAG2. Moreover, this reconstituted cRAG1-like protein could mediate the assembly of antigen receptor genes in RAG1-deficient mice. This research provided the first functional candidate for RAG1 ancestor outside of the jawed vertebrates.
The paper entitled “
An amphioxus RAG1-like DNA fragment encodes a functional central domain of vertebrate core RAG1” was published online in
Proc Natl Acad Sci USA on December 24, 2013. The work was supported by grants from 973 program and National Natural Science Foundation of China.
CONTACT:
LIU Xiaolong, Principle Investigator
Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences
Shanghai 200031, China.
Email:
liux@sibs.ac.cn
The structural and functional analyses of bfRAG1L. (A) Alignment between RAG1 and bfRAG1L. (B) The predicted secondary structure is labeled with cylinders (α-helices) and arrows (β-strands). (C) Sequences alignment of the conserved nuclease active motif in bfRAG1L, several herpes virus nucleases and EcoR V. The nuclease activities of bfRAG1L on the substrates (D) plasmid pBR322, (E) genomic DNA or (G) RNA from HEK-293T cells are shown. (F) The re-ligation products of BamH I digested linear plasmid pBR322 degraded by bfRAG1L or DNase I. (Image by Dr. LIU Xiaolong’s group)
 Appendix:
Appendix: