Pathogenic Mitochondrial tRNA Mutations Induce Multilevel Defects
Source:
Time: 2015-09-20
The human mitochondrial tRNAs (hmtRNAs) are less thermodynamically stable as they generally contain more mismatches and A/U base pairs than those in bacterial and cytoplasm. Point mutations in hmtRNAs cause various disorders such as Chronic Progressive External Ophthalmoplegia (CPEO) and Mitochondrial Myopathy (MM). Now researchers from Chinese Academy of Sciences unravel two substitutions found in the TΨC-arms of two hmtRNAsLeu isoacceptors would induce multilevel functional and structural defects.
Mitochondrial tRNALeu, especially UUR isoacceptor is recognised as a hot spot for pathogenic mitochondrial DNA point mutations. So far, 40 mutations have been reported in hmtRNAsLeu. WANG Meng and his colleagues, led by Professor WANG Enduo in Shanghai Institute of Biochemistry and Cell Biology, described the impacts of two substitutions found in the TΨC-arms of two hmtRNAsLeu isoacceptors. The G52A substitution, corresponding to the pathogenic G12315A mutation in tRNALeu (CUN) and G3283A in tRNALeu (UUR), exhibited structural changes in the outer corner of the tRNA shape as shown by RNase probing. The mutation also induced reductions in aminoacylation, 3’-end processing and base modification processes. The main effects of the A57G substitution, corresponding to mutations A12320G in tRNALeu(CUN) and A3288G in tRNALeu(UUR), was observed on the aminoacylation activity and binding to human mitochondrial Elongation Factor Tu (hmEF-Tu).
This study showed G52A and A57G mutations in the D-arm domain induced multiple effects on tRNA metabolism. They influenced not only aminoacylation but also the ability of the tRNA to be a substrate for processing or modification enzymes or to function in subsequent steps of protein synthesis. As G52A and A57G mutations widely impact on different levels of the translation process, a treatment strategy for mitochondrial diseases to be considered, such as importing mitochondrially targeted functional aminoacyl-tRNA synthetase or tRNAs.
AUTHOR CONTACT:
WANG Enduo
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
Phone: 86-21-54921241;
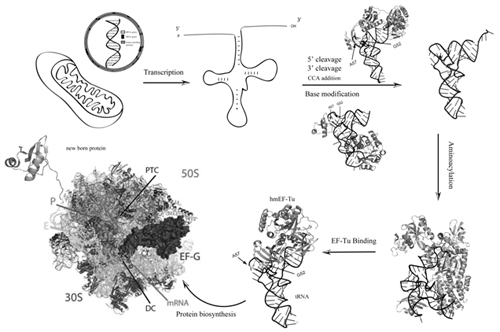
Lifecycle of hmtRNAs, including transcription, 5’- and 3’-end processing of precursor tRNA, post-transcriptional modification of bases in tRNA maturation, aminoacylation and formation of tRNA-ribosomal complexes in protein biosynthesis. (Image provided by Prof. WANG Enduo’s lab)
 Appendix:
Appendix: