Telomeres, the ends of linear eukaryotic chromosomes, are highly specialized structures that are essential for genome integrity and stability. In most eukaryotes telomere length is replenished by telomerase, a specialized reverse transcriptase that iteratively adds telomeric repeats at the chromosome ends. In Tetrahymena thermophila, TERT, TER, and p65 form the catalytic core of telomerase. In addition to this catalytic core, three protein factors p75, p45, and p19 form a subcomplex in the telomerase holoenzyme. Depletion of any of these proteins results in telomere shorting, suggesting that 7-4-1 plays an important role in telomere homeostasis. However, the primary sequences of p75, p45, and p19 lack evident homology with other proteins, and the mechanism of their functions at telomeres remains poorly understood.
WAN Bingbing and his colleagues from a research group led by Prof. LEI Ming at National Center for Protein Science Shanghai, Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, present the crystal structures of p19, the C-terminal domain of p45, and the p45N-p19 complex. Strikingly, all the structures clearly indicate that p45 and p19 are the Tetrahymena homologs of Stn1 and Ten1, two components of the CST (Cdc13/Ctc1-Stn1-Ten1) complex. CST complex is essential for telomere end formation and protection and is conserved in yeast, plants, and mammals, but has not been identified in Tetrahymena. Similar to CST complexes, 7-4-1 forms a stable heterotrimeric complex and specifically binds to telomeric single-stranded DNA. Furthermore, overexpression of wild-type p45 and p19 induces drastic telomere shortening, while overexpression of p45-binding deficient mutant of p19 does not change C-strand length but causes over-elongation of the G-strand without a matching amount of C-strand elongation. All the results reveal that telomerase holoenzyme 7-4-1 subcomplex is the Tetrahymena CST and plays a unique role in coordinating telomere G-strand and C-strand synthesis.
This study entitled “The Tetrahymena telomerase p75–p45–p19 subcomplex is a unique CST complex” was published online in Nature Structural and Molecular Biology on November 9, 2015. This work was supported by grants from the Ministry of Science and Technology of China, the National Natural Science Foundation of China, and the Strategic Priority Research Program of the Chinese Academy of Sciences.
AUTHOR CONTACT:
WU Jian and LEI Ming
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
Shanghai 200031, China.
E-mail: wujian@sibs.ac.cn, leim@sibcb.ac.cn.
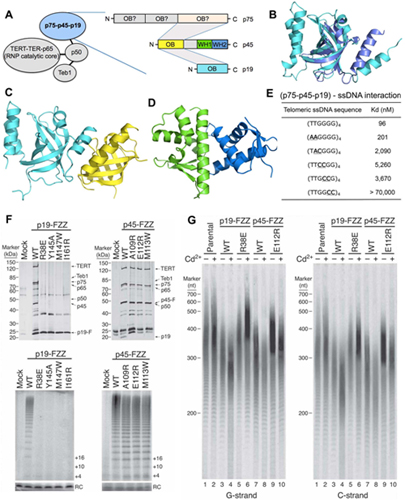
Figure. Strucutural and functional analysis of p75-p45-p19. (A) Components of Tetrahymena telomerase holoenzyme and domain organization of p75, p45 and p19. The shaded areas indicate domain interactions among p75, p45 and p19. (B) Superposition of p19 and fission yeast (Sp) Ten1. (C) Ribbon diagram of the p45N-p19 complex. (D) Ribbon diagram of C-terminal WH motifs of p45. (E) In vitro MST binding data of interactions between p75-p45-p19 and wild-type and mutant telomeric ssDNAs. (F) Affinity purifications from equivalent amounts of cells overexpressing the indicated proteins, assayed by SDS-PAGE and silver staining (upper panel) or direct primer-extension assay for telomerase activity (lower panel). (G) Denaturing gel electrophoresis and Southern blotting detecting G-strand (left panel) and C-strand (right panel) telomeric
repeat-tract lengths in cell lines overexpressing
the indicated protein or in the parental CU522 cell line.
 Appendix:
Appendix: