Histone methylation is a critical chemical modification for epigenetic regulation and plays important roles in multiple cellular events, including transcription regulation, heterochromatin formation, X-chromosome silencing. Abnormal histone methylation has been linked to many human diseases and cancers. Methylation of histone H3 Lys4 (H3K4), which is predominantly associated with actively transcribed genes, is mainly mediated by MLL family histone lysine methyltransferases (HKMTs). Mammalian MLL family HKMTs contain six members (MLL1–MLL4, SET1A and SET1B), each of which has crucial yet non-redundant roles in cells. MLL1 has been the most intensively studied because of its involvement by chromosomal translocations in a variety of acute lymphoid and myeloid leukaemias. In contrast to most SET [SU(VAR)3-9, E(Z) and TRX]-domain-containing methyltransferases, the histone lysine methyltransferase (HKMT) activity of MLL1 by itself is severely comprised, which is stimulated by three conserved factors, WDR5, RbBP5, and Ash2L shared by all MLL-family complexes. However, the molecular mechanism of activity regulation of MLL1 remains elusive, and whether other MLL proteins are regulated by these three components in the same way as MLL1 is totally unknown.
LI Yanjing and her colleagues from the research groups led by Prof. LEI Ming and Prof. CHEN Yong at National Center for Protein Science Shanghai, Institute of Biochemistry and Cell Biology, and Prof. LI Guohui from Dalian Institute of Chemical Physics, Chinese Academy of Sciences, discover that a minimized RbBP5-Ash2L heterodimer is the structural unit to interact with and activate all MLL-family histone methyltransferases. Their structural, biochemical, and computational analyses reveal a two-step activation mechanism of MLL-family proteins: interaction with the RbBP5-Ash2L heterodimer reduces the inherent flexibility of MLLSET and favors to form a catalytically competent conformation; and then H3 substrate binding induces a local conformational change in the SET-I motif of MLLSET to achieve the fully active configuration that facilitates methyl transfer process. These findings provide unprecedented insights into the common theme and functional plasticity in complex assembly and activity regulation of MLL-family methyltransferases, and also suggest a universal regulation mechanism for most histone methyltransferases.
This study entitled “Structural basis for activity regulation of MLL family methyltransferases” was published online in Nature on Feb 17, 2016. Yanjing Li from Prof. CHEN Yong's lab is the first author of this paper. Prof. LEI Ming, Prof. CHEN Yong and Prof. LI Guohui are co-corresponding authors of this paper. This work was collaborated with Dr. DOU Yali from University of Michigan, Dr. ZHANG Jian from Shanghai Jiaotong University, Dr. TIAN Changlin from University of Science and Technology of China and Dr. LI Dangsheng from Shanghai Information Center for Life Sciences. This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences, the Ministry of Science and Technology of China, the National Science and Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ of China, the National Natural Science Foundation of China, and the Basic Research Project of Shanghai Science and Technology Commission.
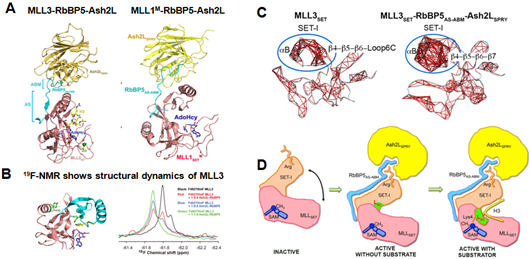
Figure. Molecular mechanism for activity regulation of MLL-family methyltransferases. (A) The crystal structure of MLL3/MLL1M-RbBP5-Ash2L in complex with cofactor product AdoHcy and the H3 peptide. (B) One-dimensional 19F-NMR measurements of MLL3SET with substitution of F4827tfmF (top) in the absence or presence of RbBP5-Ash2L. (C) The most highly correlated residues (correlation coefficients greater than 0.55) of SET-I in MD simulation are indicated by red lines. (D) A working model for the activation of MLL-family methyltransferases: interaction with the RbBP5-Ash2L heterodimer reduces the inherent flexibility of MLLSET and favors to form a catalytically competent conformation; and then H3 substrate binding induces a local conformational change in the SET-I motif of MLLSET to achieve the fully active configuration that facilitates methyl transfer process.
 Appendix:
Appendix: