Published as a full article in Cell Reports on Apr. 19th, 2016, a research team led by Prof. CHEN Lingling, at Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences(CAS), in collaboration with Prof. YANG Li, at CAS-MPG Partner Institute for Computational Biology (PICB), studied the link between circular RNAs (circRNAs) processing and transcription using 4sUDRB-seq. They found that circRNA production from pre-mRNA back-splicing is slow and largely occurs post-transcriptionally. The authors argue that circRNAs that are abundant at a steady-state level tend to be transcribed quickly and accumulate.
CircRNAs are produced from pre-mRNA back-splicing. During back-splicing, a downstream 5’ splice site is joined to an upstream 3’ splice site in a reversed orientation, resulting in a circular RNA molecule with a 3’,5’ phosphodiester bond at the back-splicing junction site. Although the majority of circRNAs still lack functional annotation, recent observations are beginning to reveal that circRNAs may play potentially important roles in gene regulation. For example, a few abundant circRNAs could act as miRNA sponges, and a set of intron-containing circRNAs was shown to regulate RNA polymerase II (Pol II) transcription. In addition, circRNAs may play important physiological roles in different biological processes. Many circRNAs are upregulated during neurogenesis, indicating that certain cricRNAs may be functional involved in neurogenesis. Both cis-complementary sequences and trans-factors are required to promote back-splicing by bringing the downstream donor and upstream acceptor sites close together. Understanding how circRNAs themselves are regulated is important to delineate their associated function.
The authors studied circRNA processing using metabolic tagging of nascent RNAs with 4-thiouridine (4sU) in human embryonic carcinoma line PA1 cells, human embryonic stem cell H9 line and H9-differentiated Forebrain neurons. The authors found that the efficiency of circRNA processing from pre-mRNA is extremely low endogenously. Back-splicing outcomes correlate with fast RNA Polymerase II elongation rate and are tightly controlled by cis –elements in vivo. For example, when the intronic RNA pairing across the circle-forming exons was disrupted by CRISPR-Cas9 mediated genome editing, increased transcription elongation rate could no longer induce any circRNA formation. Additionally, prolonged 4sU labeling in cells shows that circRNAs are largely processed post-transcriptionally and that some circRNAs are extremely stable. Circular RNAs that are abundant at a steady-state level tend to accumulate. This is particularly true in cells, such as neurons, that have slow division rates. Together, this study uncovers previously unknown features of circRNA biogenesis by investigating the link between nascent circRNA processing and transcription.
This work was supported by the National Natural Science Foundation of China, Ministry of Science and Technology of China, and Chinese Academy of Sciences.
CONTACT:
CHEN Lingling
Shanghai Institute of Biochemistry and Cell Biology (SIBCB), Chinese Academy of Sciences, Shanghai, China
Email: linglingchen@sibcb.ac.cn
Tel: +86-21-54921021
Article Link: http://dx.doi.org/10.1016/j.celrep.2016.03.058.
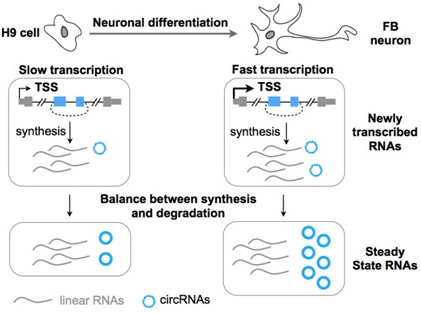
A combined effect results in the abundant but dynamic expression of circRNAs upon neuronal differentiation. On one hand, high expression of circRNAs in neurons is associated with enhanced transcription of their parental genes (top). On the other hand, upon neuronal differentiation, circRNAs accumulate post-transcriptionally to high levels while their linear counterparts do not (bottom). (Image provided by Prof. CHEN Lingling 's lab)
 Appendix:
Appendix: