Complex systems in biology often undergo slow changes affected by various external factors, whose persistent effects sometimes result in drastic or qualitative changes of system states from one stable state (i.e., the before-transition state) to another stable state (i.e., the after-transition state) through a pre-transition state or critical state. Identifying this critical state just before the occurrence of a phase transition is a challenging task, because the state of the system may show little apparent change before this critical transition during the gradual parameter variations. Such dynamics of the phase transition is generally composed of three stages, i.e., before-transition state, pre-transition state (or critical state), and after-transition state, which can be considered as three different Markov processes.
To address this issue, Prof. CHEN Luonan and his group from Key Laboratory of Systems Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, collaborating with the group of Prof. LIU Rui in South China University of Technology, developed a new method for identifying early-warning signals of critical transitions by hidden Markov model (HMM). Specifically, in this method they designed a model-free computational strategy by exploring the rich dynamical information provided by high-throughput data. They focused on detecting the switching point of the two Markov processes from the before-transition state (a stationary Markov process) to the pre-transition state (a time-varying Markov process), thereby identifying the pre-transition state or early-warning signals of the phase transition. The effectiveness of their method has been validated by detecting the signals of the imminent phase transitions of complex systems based on the simulated datasets and real datasets including the acute lung injury triggered by phosgene inhalation, MCF-7 human breast cancer caused by heregulin, and HCV-induced dysplasia and hepatocellular carcinoma. Both functional and pathway enrichment analyses validate the computational results.
This work entitled “Detecting critical state before phase transition of complex biological systems by hidden Markov model” was published in Bioinformatics on March 19, 2016.
This work was supported by the grants from the Chinese Academy of Sciences and the National Natural Science Foundation of China.
CONTACT:
CHEN Luonan;
Key Laboratory of Systems Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
Shanghai 200031, China.
E-mail: lnchen@sibs.ac.cn
Keywords: Dynamical network marker; hidden Markov model; critical transition;
Link to publication:
http://bioinformatics.oxfordjournals.org/content/early/2016/03/18/bioinformatics.btw154.full.pdf+html?sid=6d437610-d19b-4acd-8133-5c90d753265e
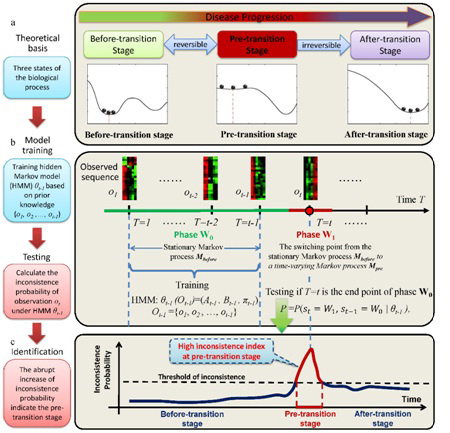
Outline for identifying the pre-transition state by using Markov model. (a) A complex biological process can be divided into three states, i.e., the before-transition state, the pre-transition state, and the after-transition state. Both the before-transition and after-transition states are relatively stable or stationary with high resilience or low fluctuations, insensitive to the parameter changes. (b) First, based on the observed sequence of samples (Ot-1={o1, o2, ..., ot-1}), we train the hidden Markov model (HMM) , that is, in view of stable dynamics, the before-transition state is modelled as the stationary Markov process , while on the other hand, the pre-transition state is defined as a time-varying Markov process, due to the dynamical characteristics that the pre-transition state has strong fluctuations and is also sensitive to parameter changes. (c) The large value of P indicates that a candidate point T=t is the switching point of with high probability. (Image provided by Prof. CHEN Luonan’s group)
 Appendix:
Appendix: