Quantitatively identifying direct dependencies between variables is an important task in data analysis, in particular for reconstructing various types of networks and causal relations in science and engineering. One of the most widely used criteria is partial correlation, but it can only measure linearly direct association and miss nonlinear associations. However, based on conditional independence, conditional mutual information (CMI) is able to quantify nonlinearly direct relationships among variables from the observed data, superior to linear measures, but suffers from a serious problem of underestimation, in particular for those variables with tight associations in a network, which severely limits its applications.
To address this issue, Prof.CHEN Luonan and his group for Key Laboratory of Systems Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy Sciences, proposed a new concept, “partial independence,” with a new measure,“part mutual information” (PMI), which not only can overcome the problem of CMI but also retains the quantification properties of both mutual information (MI) and CMI. Specifically, they first defined PMI to measure nonlinearly direct dependencies between variables and then derived its relations with MI and CMI. Finally, they used a number of simulated data as benchmark examples to numerically demonstrate PMI features and further real gene expression data from Escherichia coli and yeast to reconstruct gene regulatory networks, which all validated the advantages of PMI for accurately quantifying nonlinearly direct associations in networks.
This work entitled “Part mutual information for quantifying direct associations in networks” was published in Proceedings of the National Academy of Sciences of the United States of America on April 18, 2016.
This work was supported by the grants from the Chinese Academy of Sciences and the National Natural Science Fundation of China.
CONTACT
CHEN Luonan
Key Laboratory of Systems Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy Sciences
Shanghai 200031, China
E-mail: lnchen@sibs.ac.cn
Link: http://www.pnas.org/content/early/2016/04/15/1522586113.abstract
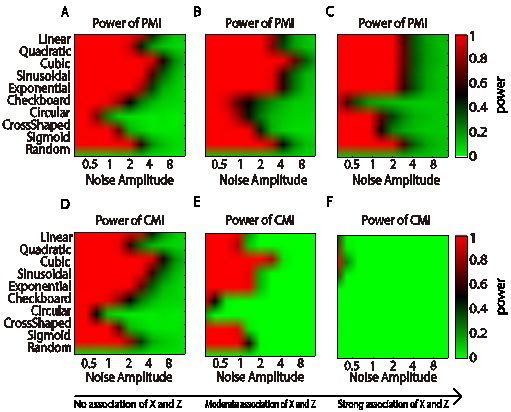
Comparison of statistical power between PMI(X;Y|Z) and CMI(X;Y|Z). Ten relationships between X and Y were used Y = f(X,Z) + a η, where “Random” means X and Y have no direct association, η is normally distributed noise with mean 0 and SD 1, and a is noise amplitudes ranging from 0 to 8. A and D show the power of PMI and CMI when X and Z are independent, but X and Y are both related with Y; B and E are the results when X is associated with Z moderately. C and F are the cases when X are strongly associated with Z, which means X is almost a copy of Z. (Image provided by Prof. CHEN Luonan's lab)
 Appendix:
Appendix: