Compared with qRT-PCR and microarray approaches designed for detecting known small RNAs, deep sequencing provides an opportunity to identify unknown ones, as well as to analyze the small RNAs with extremely diverse sequences, nucleotide editing and sequence variations at the 5’ or 3’ ends. However, due to the technical requirement of several hundred nanograms (ng) of total RNA, corresponding to thousands of oocytes or zygotes that are not readily obtainable, very few analyses have been conducted using deep sequencing to profile small RNAs in the early embryos of mammals due to RNA scarcity, which impedes investigations of the expression dynamics and modifications of small RNAs, as well as of their functions.
On June 10th, 2016, a team of researchers, led by Prof. WU Ligang of National Center for Protein Science·Shanghai, Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Sciences (SIBS), Chinese Academy of Sciences(CAS), has reported an improved small RNA sequencing method requiring only 10 ng of total RNA and revealed dynamic modifications and activities of small RNAs during mouse fertilization and early embryonic development. This work was published in Science Advances.
The researchers have optimized the method for constructing a small RNAs cDNA library such that it only requires less than 50 oocytes or 10 ng total RNA as input and can detect small RNAs with a broad abundance range of >10,000-fold. Taking advantage of this more sensitive deep sequencing method, they profiled small RNAs in mouse oocytes and 1C-8C embryos and found that the first wave of miRNA production can be detected in the zygotes as early as about 5 hours after fertilization. Although highly expressed in sperm, the small RNAs carried by a single sperm were extremely low when compared with the ones already existing in an oocyte.
Moreover, they observed an extensive degradation of maternal miRNAs at the 1C and 2C stages, which coincides with the temporal rising of miRNA adenylation during embryonic development. Interestingly, only half of the miRNA species are adenylated, and they decay slower than the non-adenylatable ones, suggesting that adenylation may protect miRNAs from massive maternal miRNA degradation in maternal-zygotic transition (MZT) when maternally inherited RNAs being rapidly degraded and zygotic genes being extensively activated, resulting in a rapid cell fate transition from gametic to totipotent.
Next, by analyzing the small RNA sequencing data together with the previous large-scale transcriptome data, they found that the function of miRNA is suppressed in 1C-2C and switched on again after the 2C stage in the mouse embryos. In contrast to non-conserved miRNAs, many highly expressed conserved maternal miRNAs are actively expressed after fertilization and target for degradation nascent mRNAs transcribed during the early wave of gene activation, as well as many maternal genes, such as Lin28 and Tet3,suggesting that, in addition to their roles in oogenesis, maternal miRNAs appear to have important functions in early embryonic development. These miRNAs are stored in the mature oocyte with their activity repressed and are then reactivated after the 2C stage to regulate early zygotic genes post-transcriptionally along with their newly produced siblings.
Their study thus provided valuable data sets for further examination of small RNAs in early embryos, and more importantly, a robust and sensitive small RNA profiling method that can be applied to other studies where RNA material is scarce.
This work was conducted in collaboration with Prof. SHI Huijuan’s lab from Shanghai Institute of Planned Parenthood Research.
This work entitled “Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos” has been published online in Science Advances on June 10th, 2016.
This study was supported by the grants from the Ministry of Science and Technology and National Natural Science Foundation of China.
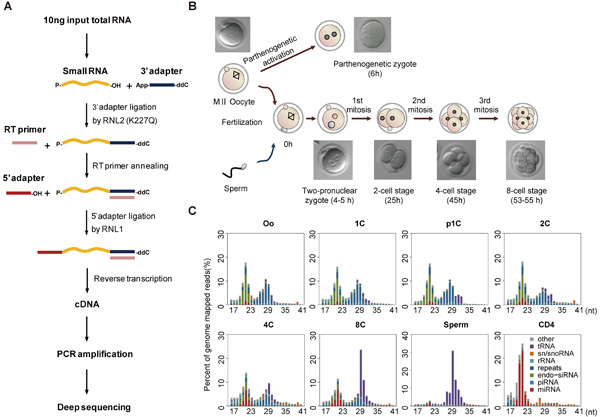
Figure. (A) Diagram of the workflow for constructing the cDNA library of small RNAs for deep sequencing. (B) Collected mouse oocyte and embryo samples at different developmental stages for small RNA sequencing. (C) Composition of small RNA categories according to their length distributions in each sample. Mouse CD4+ T cell (CD4) sample serves as a somatic cell control. (Image provided by Prof. WU Ligang’s lab)
AUTHOR CONTACT:
WU Ligang, Ph.D., Professor
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
320 Yueyang Road, Shanghai 200031, China
Tel: 86-21-54921321
Fax: 86-21-54921322
Email: lgwu@sibcb.ac.cn
KEYWORDS:
small RNA; microRNA; endo-siRNA; piRNA; adenylation; tailing; oocyte; zygote; embryo; maternal-zygotic transition.
NEWS ABSTRACT:
The researchers report an improved method to prepare the cDNA libraries of small RNAs that reduces requirement of input total RNA 10 to 100 folds, requiring as little as 10 ng of total RNA or 50 oocytes. Using this highly sensitive method, they profiled small RNAs in mouse oocytes and 1-cell to 8-cell embryos and unveiled dynamic regulation of small RNA expression in early embryonic development. Remarkably, they found that the function of miRNAs appears to be suppressed from the oocyte to 1-cell stages but is reactivated after the 2-cell stage to regulate genes that are important for development. Therefore, this study presents valuable data sets for further examination of small RNAs in early embryos, and more importantly, a robust and sensitive small RNA profiling method that can be applied to other studies where RNA material is scarce.
 Appendix:
Appendix: