A research team led by Prof. CHEN Lingling, at Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Sciences (SIBS), Chinese Academy of Sciences(CAS), annotated different types of alternative back-splicing and alternative splicing events in circRNAs. This work was published in Genome Research.
In 2014, Prof. CHEN Lingling’s group and collaborators demonstrate the mechanism of circular RNA biogenesis and present the generality of alternative circularization for the complexity of mammalian posttranscriptional regulation. Alternative formation of RNA pairing and the competition between them lead to alternative circularization, resulting in multiple circular RNA transcripts produced from a single gene.
In this study, by using the upgraded CIRCexplorer2, the authors presented a detailed map of alternative back-splicing/splicing in circRNAs,. Similar to linear message RNAs, all four basic types of alternative splicing also have been identified within circRNAs. Interestingly, many previously unannoted exons were predominantly detected in circRNAs by the de novo assembly. In addition, competition of paired intronic complementary sequences was further demonstrated to lead to alternative back-splice site selection. The annotation of alternative back-splicing and alternative splicing in circRNAs further expands the understanding of the complexity of circRNA biogenesis, and also provides a valuable resource for studying the potential functions of circRNAs.
The study entitled “Diverse alternative back-splicing and alternative splicing landscape of circular RNAs” has been published in Genome Research on June 30th, 2016.
This work was conducted in collaboration with Prof. YANG Li, at CAS-MPG Partner Institute for Computational Biology (PICB), Shanghai Institutes for Biological Sciences (SIBS), Chinese Academy of Sciences (CAS).
This work was supported by the National Natural Science Foundation of China, Ministry of Science and Technology of China, and Chinese Academy of Sciences.
CONTACT:
CHEN Lingling
Shanghai Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
Email: linglingchen@sibcb.ac.cn
Tel: +86-21-54921021
KEYWORDS: alternative back-splicing, alternative splicing, circular RNAs, complementary sequence, cassette exon
NEWS ABSTRACT: Researchers reveal the diverse alternative (back-) splicing in back-spliced circular RNAs (circRNAs). The combination of alternative back-splicing and alternative splicing robustly increases the amount of cirRNAs. The identification of different types of alternative back-splicing and alternative splicing in circRNAs from various cell lines expands the understanding of gene expression regulation and extends the complexity of mammalian posttranscriptional regulation.
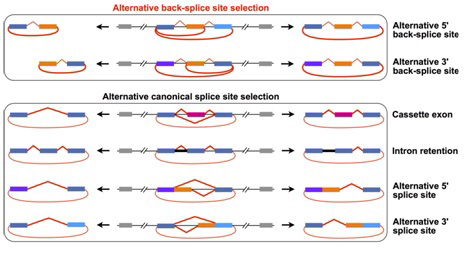
An upgraded computational pipeline (CIRCexplorer2) systematically identifies alternative (back-) splicing in back-spliced circular RNAs (circRNAs). (Image by CHEN Lingling’s research group)
 Appendix:
Appendix: