Methylation at the N6 position of adenosine (m6A) is the most abundant internal modification within protein-coding and long noncoding RNAs in eukaryotes, which is a reversible process with important biological functions. YT521-B homology domain family (YTHDF) proteins are the ¢readers¢ of m6A, the binding of which results in the alteration of the translation efficiency and stability of m6A-containing RNAs. However, the mechanism underlay is poorly understood.
A team of researchers, led by Prof. WU Ligang of National Center for Protein Science·Shanghai, Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Sciences (SIBS), Chinese Academy of Sciences (CAS), has uncovered the mechanism of YTHDF2-mediated degradation of m6A-containing RNAs in mammalian cells. This work has been published in Nature Communications.
The researchers used a pulse-chase system to monitor the decay of RNAs. They found that both m6A modification and binding of YTHDF2 can destabilized RNA by hastening deadenylation (shortening of the poly(A) tail) as the initial step. Next they screened for interactions between YTHDF2 and components of the potential cellular deadenylase complexes. The results drew their attention to CCR4-NOT, a nine-subunit complex containing two deadenylase subunits (CAF1 and CCR4) and a large scaffold subunit, CNOT1. Dominant-negative assay confirmed that CAF1 and CCR4 are the ribonucleases responsible for the YTHDF2-mediated deadenylation. Further study showed that YTHDF2 recruited the CCR4-NOT complex through a direct interaction between the YTHDF2 N-terminal region and the SH domain of the CNOT1 subunit and that this recruitment was essential for the deadenylation and decay of m6A-containing RNAs.
This study uncovered the mechanism of YTHDF2-mediated degradation of m6A-containing RNAs in mammalian cells, providing a further understanding of RNA modifications on regulation of gene expression.
This work was conducted in collaboration with Prof. MA Jinbiao’s lab from School of Life Sciences, Fudan University.
This work entitled “YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex” has been published online in Nature Communications on August 25, 2016.
This study was supported by the grants from the Ministry of Science and Technology, National Natural Science Foundation of China and China Postdoctoral Science Foundation.
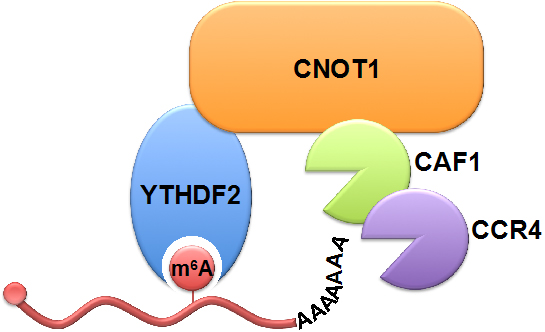
YTHDF2 binds to the m6A modification on RNA and recruits the CCR4-NOT deadenylase complex through direct interaction with CNOT1, thus promoting the deadenylation and decay of RNA. (Image provided by Prof. WU Ligang's lab)
AUTHOR CONTACT:
WU Ligang, Ph.D., Professor
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
320 Yueyang Road, Shanghai 200031, China
Tel: 86-21-54921321
Fax: 86-21-54921322
Email: lgwu@sibcb.ac.cn
KEYWORDS: m6A, YTHDF2, CCR4-NOT, deadenylation, RNA decay.
NEWS ABSTRACT:
m6A is the most abundant internal modification within protein-coding and long noncoding RNAs in eukaryotes and is a reversible process with important biological functions. YTHDF2, a reader of m6A, is responsive for the degradation of its cognate RNAs, but the mechanism is unclear. In this study, researchers demonstrate that m6A-containing RNAs exhibit accelerated deadenylation that is mediated by the CCR4-NOT deadenylase complex. Further study show that YTHDF2 recruits the CCR4-NOT complex through a direct interaction between the YTHDF2 N-terminal region and the SH domain of CNOT1, the scaffolding subunit of the CCR4-NOT complex. Therefore, this study has uncovered the mechanism of YTHDF2-mediated degradation of m6A-containing RNAs in mammalian cells.
 Appendix:
Appendix: