On October 20, 2016 (Beijing time), Nature online published a research paper entitled “TET-mediated DNA demethylation controls gastrulation by regulating Lefty–Nodal signalling”, from Dr. XU Guoliang’s group at Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Sciences (SIBS), Chinese Academy of Sciences (CAS), and Dr. XIN Sun’s group at Department of Pediatrics, University of California, San Diego (UCSD). The authors found that TET-mediated DNA oxidation modulates Lefty-Nodal signaling by promoting demethylation and/or antagonizing de novo methylation at the common target genes shared with DNMT3A and DNMT3B methyltransferases.
Mammalian genomes undergo epigenetic modifications, including cytosine methylation (5mC) by DNA methyltransferases (DNMTs). Further oxidation of 5mC by the Ten-eleven-translocation (TET) family of dioxygenases may lead to demethylation. While cytosine methylation plays key roles in multiple processes such as genomic imprinting and X-chromosome inactivation, the functional significance of cytosine methylation and its further oxidation in mouse embryogenesis remains to be fully determined.
The researchers establishes the first Tet triple knockout mouse model to inactivate all three Tet genes in mice, which leads to gastrulation phenotypes, including primitive streak patterning defects in association with impaired maturation of axial mesoderm and failed specification of paraxial mesoderm, mimicking phenotypes in embryos with gain-of-function Nodal signaling. Introduction of a single mutant allele of Nodal in the Tet mutant background partially restored patterning, suggesting that hyperactive Nodal signaling contributes to the gastrulation failure of Tet mutants. Increased Nodal signaling is likely due to diminished expression of the Lefty1 and Lefty2 genes, which encode inhibitors of Nodal signaling. Moreover, reduction in Lefty gene expression is linked to elevated DNA methylation as both Lefty-Nodal signaling and normal morphogenesis are largely restored in Tet-deficient embryos when the Dnmt3a and Dnmt3b genes are disrupted. Additionally, a point mutation in Tet that specifically abolishes the dioxygenase activity causes similar morphological and molecular abnormalities as the null mutation.
These results show that TET-mediated DNA oxidation modulates Lefty-Nodal signaling by promoting demethylation in opposition to methylation by DNMT3A and DNMT3B. These findings reveal a fundamental epigenetic mechanism featuring dynamic DNA methylation and demethylation crucial to regulation of key signaling pathways in early body plan formation.
Other researchers from China and USA also contribute to this work. They are Dr. TANG Fuchou and Dr. GE Hao from Peking University, and Dr. Deborah L. Chapman from University of Pittsburgh. This work was supported by the National Science Foundation of China, the Ministry of Sciences and Technology of China and the ‘Key New Drug Creation and Manufacturing Program’ of China.
AUTHOR CONTACT:
XU Guoliang
Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China
Phone: 86-21-54921332
E-mail: glxu@sibcb.ac.cn
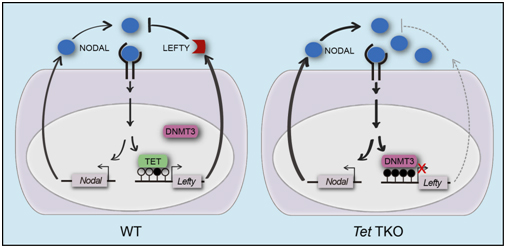
Model for the roles of TET and DNMT3 enzymes in the Lefty-Nodal feedback loops in embryonic development. (Image provided by Dr. XU Guoliang’s group)
 Appendix:
Appendix: