On Oct. 24, 2016, Nature Structural & Molecular Biology online published a research paper entitled “Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM”, from Dr. CONG Yao’s group at Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, CAS. Combining high-resolution cryo-EM and biochemical analysis, this research team revealed as taggered ATP binding mechanism of eukaryotic chaperonin TRiC, and unambiguously identified the subunit locations in open-state TRiC. Their study offers new insight into how the protein folding nanomachine TRiC prepares itself for the action of substrate productive folding.
The eukaryotic chaperonin TCP1-ring complex (TRiC) folds approximately 10% of newly synthesized cytosolic proteins and mediates the folding of many essential structural and regulatory proteins, such as actin, tubulin, CDC20, and VHL tumor suppressor, etc. Hence, dysfunction of TRiC is closely associated with cancer and neurodegenerative diseases. The hydrolysis of ATP is essential for the TRiC-meditated folding of nascent or misfolded substrate. Extensive efforts have been made to resolve 3D structures of TRiC in distinct nucleotide states. However, the structural determination of the open state TRiC remains challenging, owing to its extremely dynamic nature, conformational heterogeneity, and the particle preferred orientation problem usually associated with the open conformation of group II chaperonins. Therefore, high-resolution structural details of the open state of TRiC and of its ATP- binding-triggered conformational transition have been lacking.
In this study, this research team presents two cryo-EM structures of Saccharomyces cerevisiae TRiC in a newly identified nucleotide partially preloaded (NPP) state and in the ATP-bound state, at 4.7-Å and 4.6-Å resolution, respectively, representing the best resolved open-state TRiC structures to date. Through inner-subunit eGFP tagging, they identified the subunit locations in open-state TRiC and found that the CCT2 subunit pair forms an unexpected Z shape. ATP binding induces a dramatic conformational change on the CCT2 side, thereby suggesting that CCT2 plays an essential role in TRiC allosteric cooperativity. Moreover, their findings indicate that TRiC has evolved into a complex that is structurally divided into two sides—the CCT2 side and the CCT6 side—whose ATP binding and corresponding conformational transitions occur in a staggered manner. This work offers insight into how the TRiC nucleotide cycle coordinates with its mechanical cycle in preparing folding intermediates for further productive folding.
ZANG Yunxiang, JIN Mingliang, and WANG Huping are the co-first-authors of the paper, and Dr. CONG Yao is the corresponding author. The authors would like to thank Prof. CHIU Wah (Baylor College of Medicine) for critical discussion of this work, and Profs. ZHOU Jinqiu and ZHOU Zhaocai (Institute of Biochemistry and Cell Biology) for their generous support. This work was funded by Chinese Academy of Sciences, the National Natural Science Foundation of China and Ministry of Science and Technology of China.
Article Link: http://www.nature.com/nsmb/journal/vaop/ncurrent/full/nsmb.3309.html
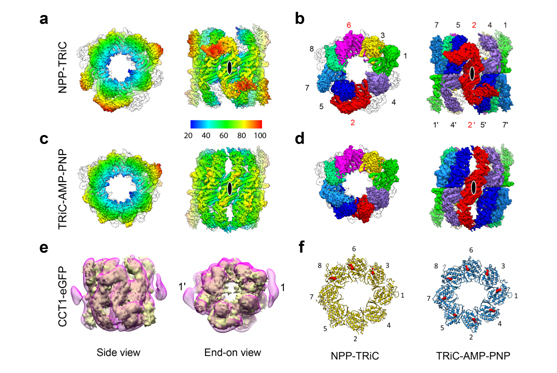
Figure legend: Cryo-EM maps of TRiC in the NPP and ATP-bound states, subunit identification through inner-subunit eGFP tagging, and distribution of nucleotide density in NPP-TRiC and TRiC–AMP-PNP.
Author Contact:
CONG Yao, Ph.D., Professor
National Center for Protein Science (Shanghai), Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
Shanghai, China, 201210
Phone: +86-21-20778199
E-mail: cong@sibcb.ac.cn
 Appendix:
Appendix: