DNA is the genetic information carrier in all cells, and its integrity and stability are essential to life. However, DNA could be attacked by either environmental factors or endogenous byproducts of metabolism, leading to mutations, genomic instability, and apoptosis. To prevent the harmful effects of DNA damage and maintain the genome integrity, all organisms have evolved various DNA repairing mechanisms to detect and repair different types of DNA lesions. In recent years, Prof. Ding’s group has focused on the proteins and protein complexes involved in DNA damage repairing pathways and has revealed the molecular mechanisms and biological functions of the MHF1-MHF2 complex (Structure, 2012) and FANCM-FAAP24 complex (Nucleic Acids Research, 2013) in the Fanconi anemia pathway.
Homologous recombination (HR) is a highly conserved error-free DNA repairing mechanism, which is responsible for repair of the most harmful DNA lesions, such as DNA double-strand breaks (DSBs) and inter-strand crosslinks (ICLs), and thus plays a critical role in maintenance of genome integrity. In recent years, a new complex named Shu complex was identified as a conserved HR regulator in most eukaryotes. In budding yeast, Shu complex consists of Csm2, Psy3, Shu1, and Shu2, and plays important roles in both DNA damage repair and tolerance via the HR pathway. So far, the structures and functions of Shu1 and Shu2 and the molecular basis for the assembly and function of the Shu complex are still unknown.
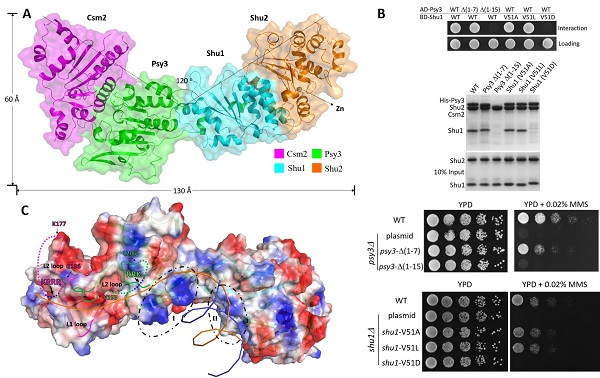
Under the supervision of Prof. DING Jianping from Institute of Biochemistry and Cell Biology (SIBCB), Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (CAS), PhD student Zhang Shicheng and his colleagues solved the crystal structure of yeast Shu complex and show that Csm2, Psy3, Shu1, and Shu2 interact with each other in sequence to form a V-shape overall structure with the Csm2-Psy3 and Shu1-Shu2 subcomplexes occupying the two arms of the “V” shape. Shu1 assumes a structure resembling the ATPase core domain of Rad51 and thus represents
a new Rad51 paralog. As the first structure of the Shu2/SWS1 family members, Shu2 assumes a novel protein fold consisting of a conserved SWIM domain and a small insertion domain. Both the SWIM domain and insertion domain have predominantly hydrophobic interactions and forms a stable subcomplex with Shu1. Analysis of the Psy3-Shu1 interface identifies two essential elements in the assembly of the Shu complex: one is the N-terminal region of Psy3 which plays a dominate role in the interaction with Shu1, and the other is Val51 of Shu1 which inserts into a hydrophobic cavity on Psy3. The key residues at the protein-protein interfaces are validated by both in vivo and in vitro functional assays. The structural and functional data reveal the molecular basis for the assembly of the Shu complex which is essential for its function in DNA damage repair. Furthermore, detailed analysis of the electrostatic surface of the Shu complex together with the previous biochemical data identifies two potential DNA-binding sites: site I is a shallow surface groove and site II consists of a flat surface and a large cleft. As the Shu complex prefers to bind forked and 3’-overhang DNAs both of which contain ssDNA and dsDNA regions, it is proposed that site I may be responsible for binding the ssDNA region of the substrate and site II may be involved in binding of the dsDNA region of the substrate, and the two binding sites work cooperatively to ensure a tight binding of the substrate. These results provide new insights into the functional role of Shu complex in DNA damage repair.
This research work entitled “Structural basis for the functional role of the Shu complex in homologous recombination” was published online in Nucleic Acids Research on Oct 24, 2017. This study was supported by grants from the National Natural Science Foundation of China, the Ministry of Science and Technology of China, and the Strategic Priority Research Program of the Chinese Academic of Sciences.
Figure legends
Structural basis for the functional role of the Shu complex in homologous recombination
(A) Overall structure of the Shu complex. (B) N-terminal region of Psy3 and Val51 of Shu1 are important for the assembly of Shu complex and its function in DNA damage
repair. (C) Possible DNA binding model for the Shu complex.
Link: https://academic.oup.com/nar/article/4563311/Structural-basis-for-the-functional-role-of-the
 Appendix:
Appendix: