On Mar. 12th, PNAS published online a work entitled “Structural insights into the sequence-specific recognition of Piwi by Drosophila Papi” from the research team of Dr. HUANG Ying from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. This work identified the direct interaction between Drosophila Piwi and Papi and provided mechanistic insights into the important role of Papi-Piwi interactions in the 3’ end trimming process of piRNA biogenesis.
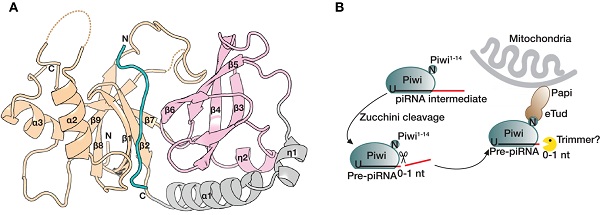
In Drosophila ovaries, the precursor piRNAs (prepiRNAs) transcribed from piRNA clusters are exported to the nuage in the cytoplasm, where the prepiRNAs are cleaved into piRNA intermediates by a mitochondrial outer membrane protein Zucchini, followed by 3’ end trimming and methylation to yield mature piRNAs. Subsequently, the mature piRNAs are loaded into PIWI proteins to form various piRISCs.
The tudor domain-containing (Tdrd) proteins, which are conserved among files, worms, and mammals, play important roles in PIWI localization and function through recognizing symmetrically dimethylated arginine (sDMA) residues of PIWI proteins by eTud domains.
Papi (partner of PIWI, also called TDRD2 or Tdrkh) is a Tdrd protein which contain an extended tudor domain (eTud). As a mitochondria outer membrane protein, Papi is proposed to recruit Piwi and trimmer to facilitate the formation of piRNA 3’ end trimming complex. In Drosophila, the depletion of papi increase the length of Piwi-bound piRNA, leaving both Aub-bound and Ago3-bound piRNAs unaffected. However, the molecular mechanism underlying the function of the Papi-Piwi complex remains unclear.
In this work, the researchers identified the region of Piwi that interacts with the eTud domain of Papi, and determined the crystal structures of Papi-eTud both in apo form and in complex with unmethylated Piwi (Piwi-unme) or symmetrically dimethylated Piwi at arginine-10 (Piwi-R10me2s). The structural and biochemical data showed, unlike the consensus eTud recognition motif which is (G/A)R repeat, Papi recognizes the RGRRR motif of Piwi in a sequence-specific manner both in vitro and in vivo. Moreover, deletion of papi results in fly fertility defects. In vivo fly rescue experiments further establish that the binding pocket of Papi is required for its function in fertility and transposon silencing. These findings shed light on the function of Papi in piRNA maturation, supporting the recent discovery that after Zuc cleavage Papi-assisted piRNA trimming is consistent with its role in silkworms and mice. This work also facilitates the identification of new PIWI-interaction partners of Tudor domain-containing proteins.
ZHANG Yuhan from SIBCB is the first author of this work. This study was supported by National Natural Science Foundation of China, Strategic Priority Research Program of the Chinese Academic of Sciences, Chinese Academy of Sciences Facility-based Open Research Program and the State Key Laboratory of Molecular Biology.
Fig. 1. (A) Overall structure of the Papi-Piwi-R10me2s complex. (B) Model for the function of Papi in the piRNA 3’ end trimming. Papi directly interacts the Piwi N terminus by the eTud domain downstream of Zuc.
 Appendix:
Appendix: