The biogenesis, functions and challenges of circular RNAs
Source:
Time: 2018-07-30
Research teams led by Dr. CHEN Ling-Ling at Shanghai Institute of Biochemistry and Cell Biology and Dr. YANG Li at CAS-MPG Partner Institute for Computational Biology, Chinese Academy of Sciences were invited to publish a review paper, entitled “The biogenesis, functions and challenges of circular RNAs”, in Molecular Cell to survey the most recent progress on circular RNA biogenesis and functions and discuss technical obstacles in circular RNA studies.
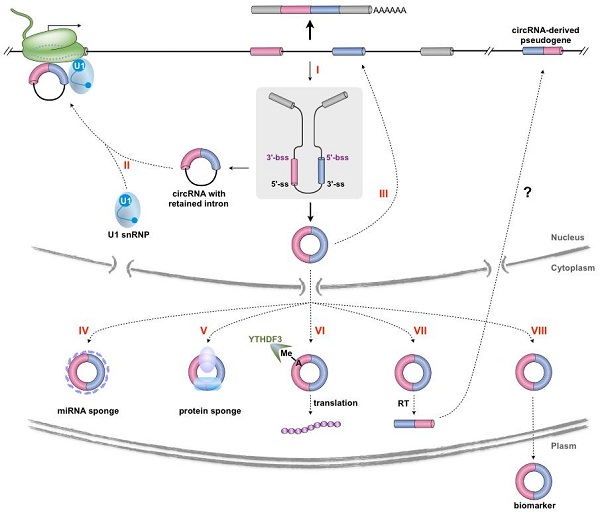
Covalently closed circular RNAs (circRNAs) are produced by precursor mRNA back-splicing of exons of thousands of genes in eukaryotes. In
general, circRNAs are expressed at low levels. However, at the individual gene level, some circRNAs are more abundant than their linear transcripts and their expression is independent of related linear isoforms. In addition, it has been found that one gene locus can produce multiple circRNAs, with mechanisms related to alternative back-splicing and alternative splicing site selection. Recent data has shown that the level of steady-state circRNA expression in cells can be regulated at three major levels, including the transcriptional regulation, modulation by
cis elements and
trans factors, as well as their turnover.
Although the generally low expression of circRNAs suggests the possibility that they are spurious members of the eukaryotic transcriptome, emerging studies have begun to reveal that at least some circRNAs play potentially important roles in physiological and pathological conditions by distinct modes of action at the molecular level, such as affecting transcription and splicing, acting through associated miRNAs or proteins as molecular sponges, being translatable to produce peptides and generating pseudogenes by retrotransposition. Nevertheless, functional implications of circRNAs have only been explored preliminarily, which is at least in part due to limitations of tools used to study them.
As sequences of individual circRNAs fully overlap with their cognate linear RNA isoforms processed from the same pre-mRNAs, dissecting the functional significance of circRNAs has been a challenge. Resolving the contribution of a circRNA from its residing gene into an observable effect remains difficult. Existing and additional potential methods that can be used to address circRNA functions and their limitations were also discussed in this comprehensive reviewer paper.
LI Xiang, a Ph.D. student in Dr. CHEN’s lab is the first author, Drs. CHEN and YANG are co-corresponding authors of this review. The YANG lab is supported by National Natural Science Foundation of China (NSFC). The CHEN lab is supported by Chinese Academy of Sciences (CAS), NSFC and Howard Hughes Medical Institute (HHMI).

 Appendix:
Appendix: