Tetraubiquitin-induced Conformations of 26S Proteasome Reveal Mechanism of Degradation Initiated by TetraUb
Source:
Time: 2019-02-26
The K48-linked polyUb chains with the length of four or more serve as the predominant proteasome-targeting signal. Structural information of tetraUb bound to proteasome is an essential piece of information to understand how proteasomes respond to a proper degradation signal.
Due to the various binding sites of polyUb chains on proteasome and the dynamic nature of proteasome itself, capturing polyUb on proteasome has been extremely challenging, and to date, no proteasome structure has resolved the molecular location of non-covalently attached polyUb. Thus, the exact molecular mechanisms underlying the substrate degradation of proteasomes initiated by polyUb remains to be elucidated.
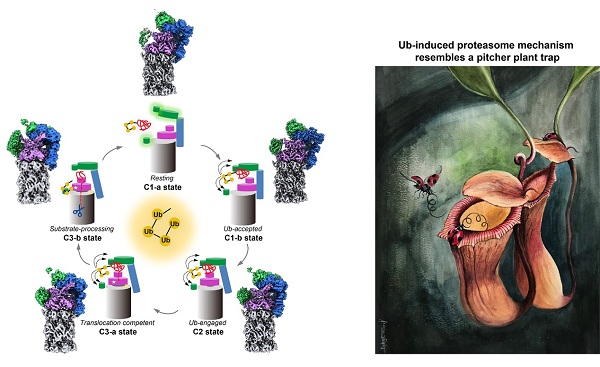
In a recent study published in Molecular Cell, Prof. CONG Yao team from Shanghai Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences resolved an ensemble of cryo-EM structures of proteasome in the presence of K48-Ub4, with three of them resolved at near-atomic resolution, localized tetraUb in an intact 26S proteasome for the first time. They also put forward a mechanistic model of a functional cycle for proteasomes induced by tetraUb.
Researchers identified a conformation with stabilized ubiquitin receptors and a previously unreported orientation of the lid, assigned as a Ub-accepted state. The upper part of RP in this state tilted ‘‘downward’’ toward Rpn1, which motion may be induced by K48-Ub4 to initiate the transition from resting to activated state.
Intriguingly, they resolved a new conformation of 26S proteasome with localized K48-Ub4 to the toroid region of Rpn1, suggesting a unique binding mode of K48-Ub4 on proteasome, and this structure was assigned as a substrate-processing state.
In collaboration with Prof. Michael H. GLICKMAN from Technion-Israel Institute of Technology, they also found the cohabitation of K48-Ub4, Rad23, and USP14/Ubp6 on Rpn1, which provides a platform for polyUb chain trimming to accelerate Ub recycling. They proposed a long-range allosteric regulation of 20S gate-opening mechanism involving the propagation of the motion of the lid to the gate region through the Rpn6-a2 interaction.
Taken together, researchers captured the location of K48-Ub4 on intact 26S proteasome for the first time, revealing a unique binding mode of K48-Ub4 on proteasome, and put forward a model of a functional cycle for proteasomes induced by tetraUb and nucleotide.
This study provided insights into the structural mechanism of proteasome allosteric coordination induced by tetraUb and took a step forward toward elucidating the structural mechanism of substrate delivery and processing by the proteasome.
The ubiquitin-proteasome system (UPS) is essential for many cellular processes, including cell cycle, regulation of gene expression, and response to proteotoxic stress. Dysfunction of the UPS is implicated in various diseases, including cancers and neurodegenerative diseases.

 Appendix:
Appendix: