A vitamin-C-derived DNA modification catalysed by an algal TET homologue
Source:
Time: 2019-05-05
5-methylcytosine (5mC) is a prevalent DNA modification found in many organisms. TET proteins iteratively oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), promoting DNA demethylation in mammalian cells. TET homologues are also present in a wide variety of other eukaryotes. However, their enzymatic activities and physiological functions remain elusive.
In a paper published in Nature on May 2nd, 2019 by a multi-site research team led by Dr. XU Guoliang from the Shanghai Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (CAS), Dr. TANG Huiru from Fudan university in Shanghai and Dr. HUANG Kaiyao from the CAS Institute of Hydrobiology in Wuhan, researchers discovered a novel vitamin C-derived DNA modification in green alga Chlamydomonas reinhardtii.
This modification is catalysed by a TET homologue, of which the catalytic mechanism and physiological function were also elucidated.
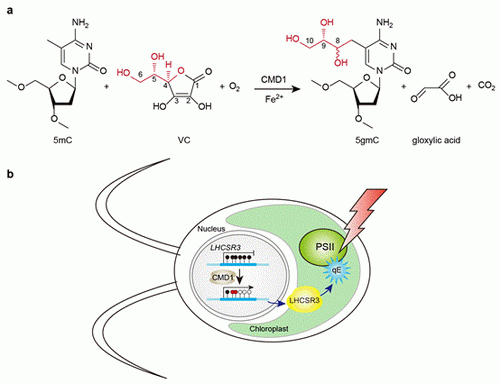
The research team firstly identified eight TET homologues by searching TET/JBP domain containing proteins in C. reinhardtii. Using 5mC-containing DNA as substrate, one of the homologues, namely CrTET1 or CMD1, is able to convert 5mC into two unidentified products.
NMR analysis showed that both products contain a glyceryl moiety at the methyl carbon of 5mC. These two stereoisomers are thus identified as 5-glyceryl-methylcytosine (5gmC).
In search of the origin of the glyceryl moiety, they found that 2-oxoglutarate, the co-substrate for the conventional dioxygenases, is dispensable for CMD1-catalysed reaction.
Surprisingly, vitamin C (VC) is an essential co-factor for CMD1 catalysis. In the reaction, the glyceryl portion of VC is transferred to 5mC to generate 5gmC in DNA, releasing CO2 and glyoxylic acid as by-products.
The VC-derived DNA modification is present in the genome of C. reinhardtii, and its level decreases significantly in a CMD1 mutant strain generated by a CRISPR/Cas9-based co-selection strategy.
Moreover, the tolerance towards high light exposure of CMD1 mutant cells is compromised compared to wild-type strains. In the absence of CMD1, LHCSR3 (Light Harvesting Complex Stress Related 3), a critical gene for protecting C. reinhardtii from photooxidative damage, is hypermethylated and downregulated compared to wild-type cells, leading to a reduced photoprotective capacity of non-photochemical quenching (NPQ), which is known to contribute to dissipate excess light energy to heat in plants.
Overall, this study uncovers a novel type of eukaryotic DNA base modification, which is generated by a divergent TET homologue and unexpectedly derived from vitamin C. This modification may counteract DNA methylation and epigenetically regulate vital physiologic processes or stress responses.
These findings highlight not only the structural versality of DNA in association of organismal environmental adaptation but also uncovers a hitherto unappreciated contribution of vitamin C in the catalytic reaction in biochemistry.
Contact: glxu@sibcb.ac.cn
Reference: https://www.nature.com/articles/s41586-019-1160-0

 Appendix:
Appendix: