Meiosis, a specialized form of cell division in most sexually reproducting organisms, serves as the basis of sexual reproduction. In a study published in Nature, researchers led by Prof. TONG Ming-Han from the Center for Excellence in Molecular Cell Science (Shanghai Institute of Biochemistry and Cell Biology) of the Chinese Academy of Sciences, in collaboration with Prof. HUANG Ying from Xinhua Hospital, Shanghai Jiaotong University School of Medicine, reports the successful in vitro reconstitution of meiotic DNA double-strand breaks (DSB) formation.
Homologous recombination—a crucial meiotic event—not only enhances genetic diversity through exchanging parental genetic substance but also establishes physical connections between homologous chromosomes to ensure their precise segregation. This process is initiated by the formation of programmed DSBs, catalyzed by Spo11, a discovery dating back to 1997.
However, despite nearly three decades of efforts, reconstituting DSB formation in vitro remained an unsolved challenge—until now. To overcome this barrier, Tang, et al. expressed and purified the SPO11-TOP6BL complex using Expi293F cells. They demonstrated that this complex cleaves DNA and covalently attaches to the 5' terminus of DNA breaks in vitro, successfully reconstituting meiotic DSB formation in vitro. Thus, this long-standing question in meiotic recombination has now been resolved.
Further investigation using a point-mutation strategy revealed that Mg2+ is essential for the complex’s DNA cleavage activity. Knock-in mice carrying a SPO11 point-mutation that disrupts Mg2+ binding exhibited a complete loss of DSB formation. Interestingly, the activity of the SPO11 complex functions independently of ATP, distinct from its ancestral enzyme, topoisomerase VI.
This study represents a major breakthrough in understanding meiotic recombination, providing a powerful platform for dissecting the molecular mechanisms of meiotic homologous recombination.
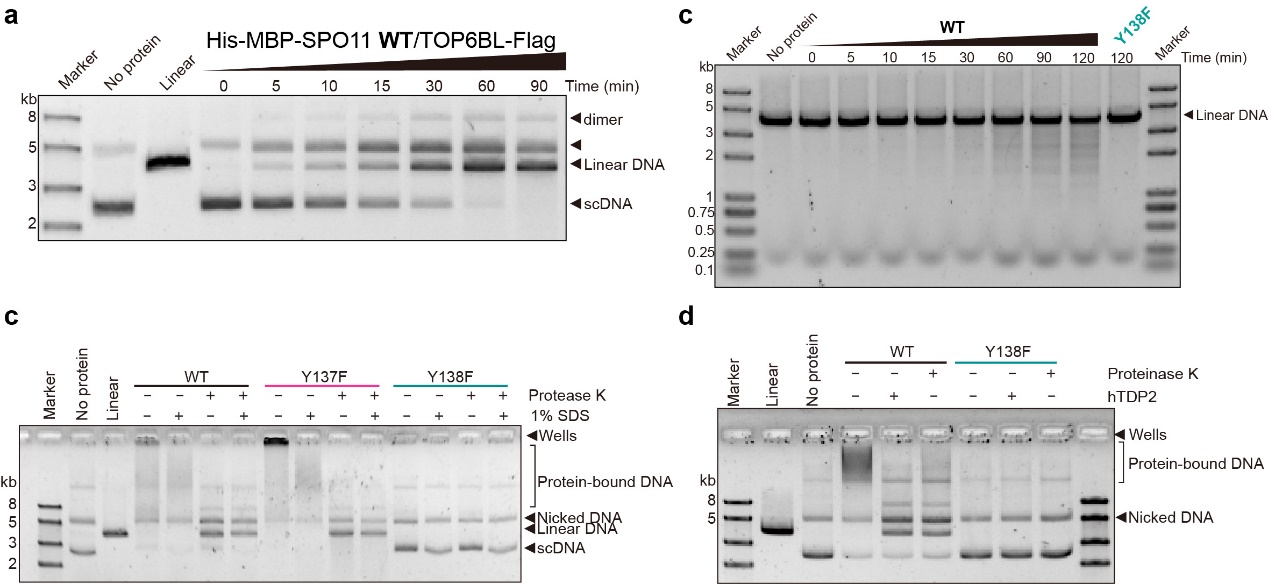
(a) The DNA cleavage experiment of SPO11-TOP6BL on supercoiled plasmid. As the reaction time increased, the plasmid was gradually cut into linear segments. (b) The DNA clevage experiment of SPO11-TOP6BL on linear DNA. As the reaction time increased, long linear DNA was gradually cleavage into DNA fragments of different lengths. (c) Adding SDS does not disrupt the interaction between protein and DNA, indicating that SPO11-TOP6BL covalently binds to DNA after cutting DNA. (d) hTDP2 can disrupt the interaction between SPO11 and DNA, indicating that the covalent connection between SPO11 and DNA is achieved through the 5' phosphotyrosine bond formed between the Y138 residue of SPO11 and the 5' end of DNA.
Reference: https://doi.org/10.1038/s41586-024-08551-1
 Appendix:
Appendix: